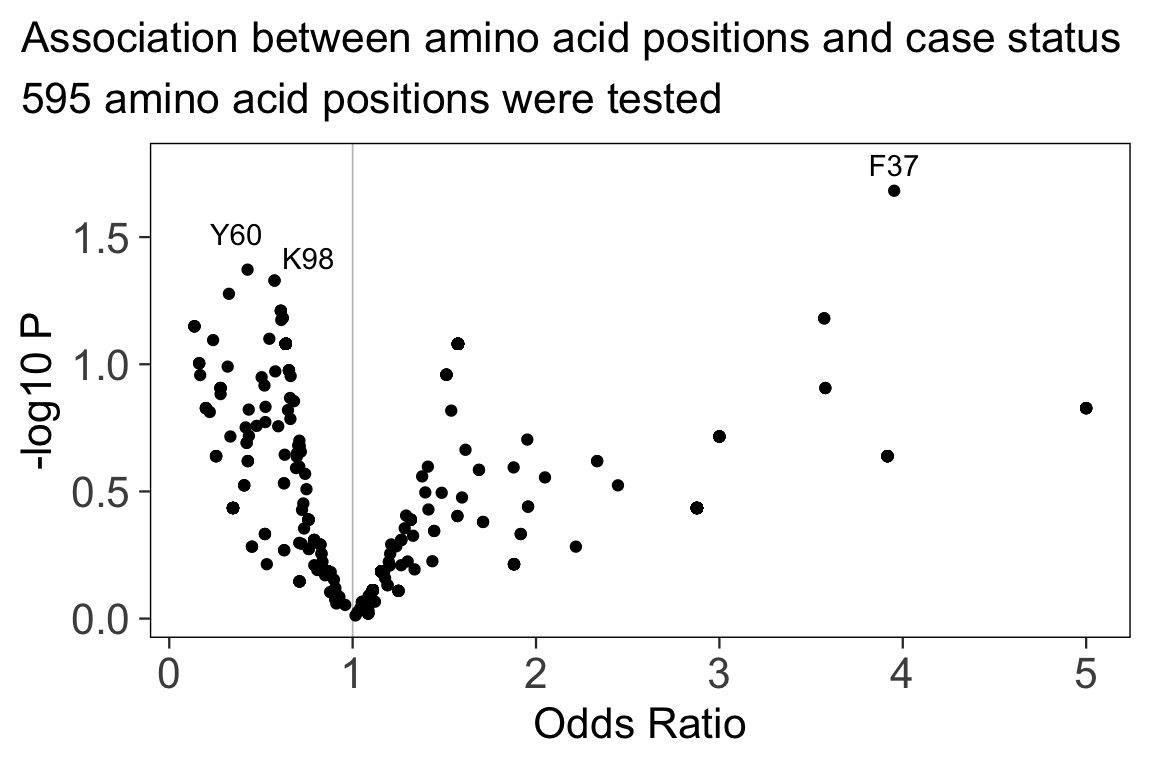
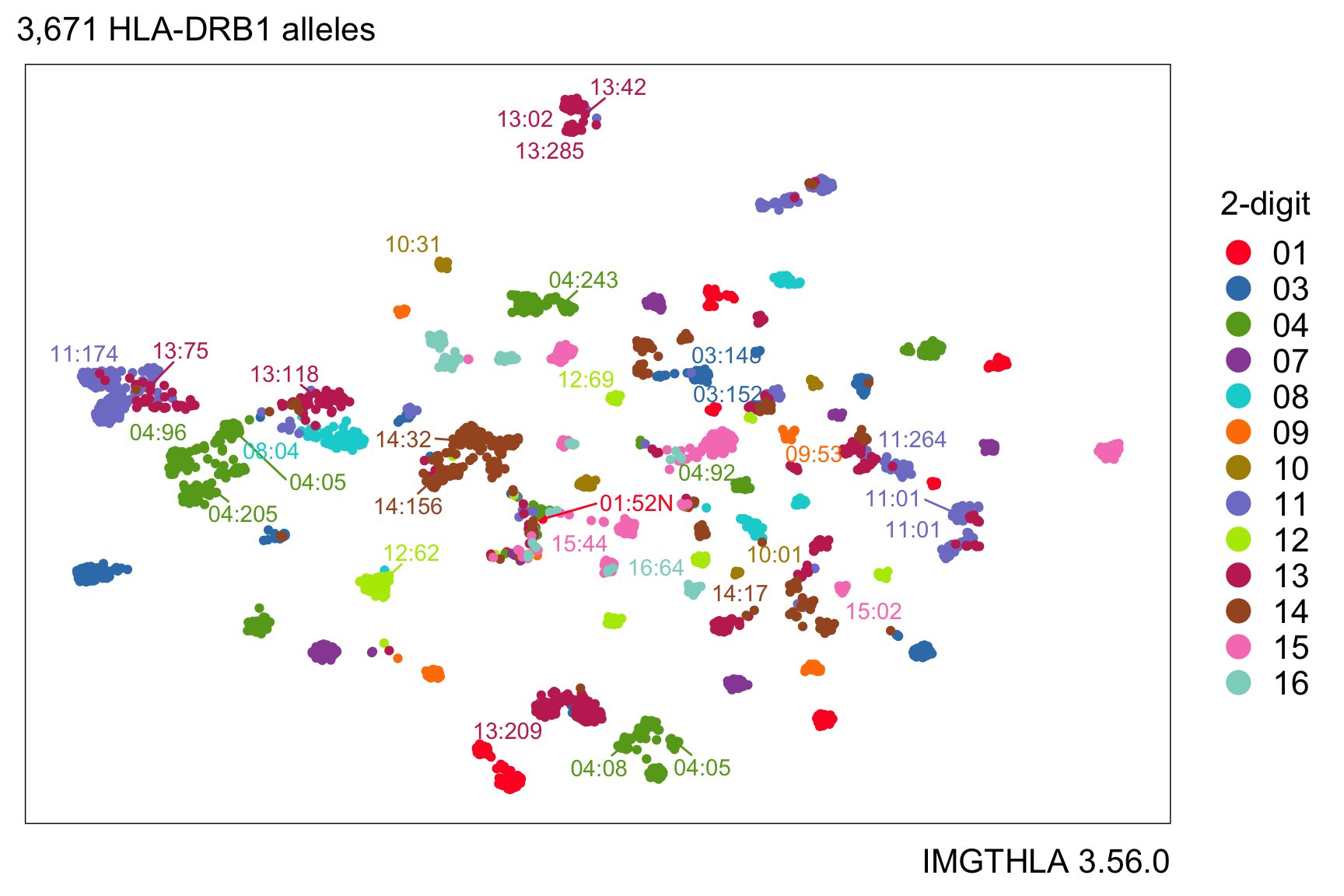
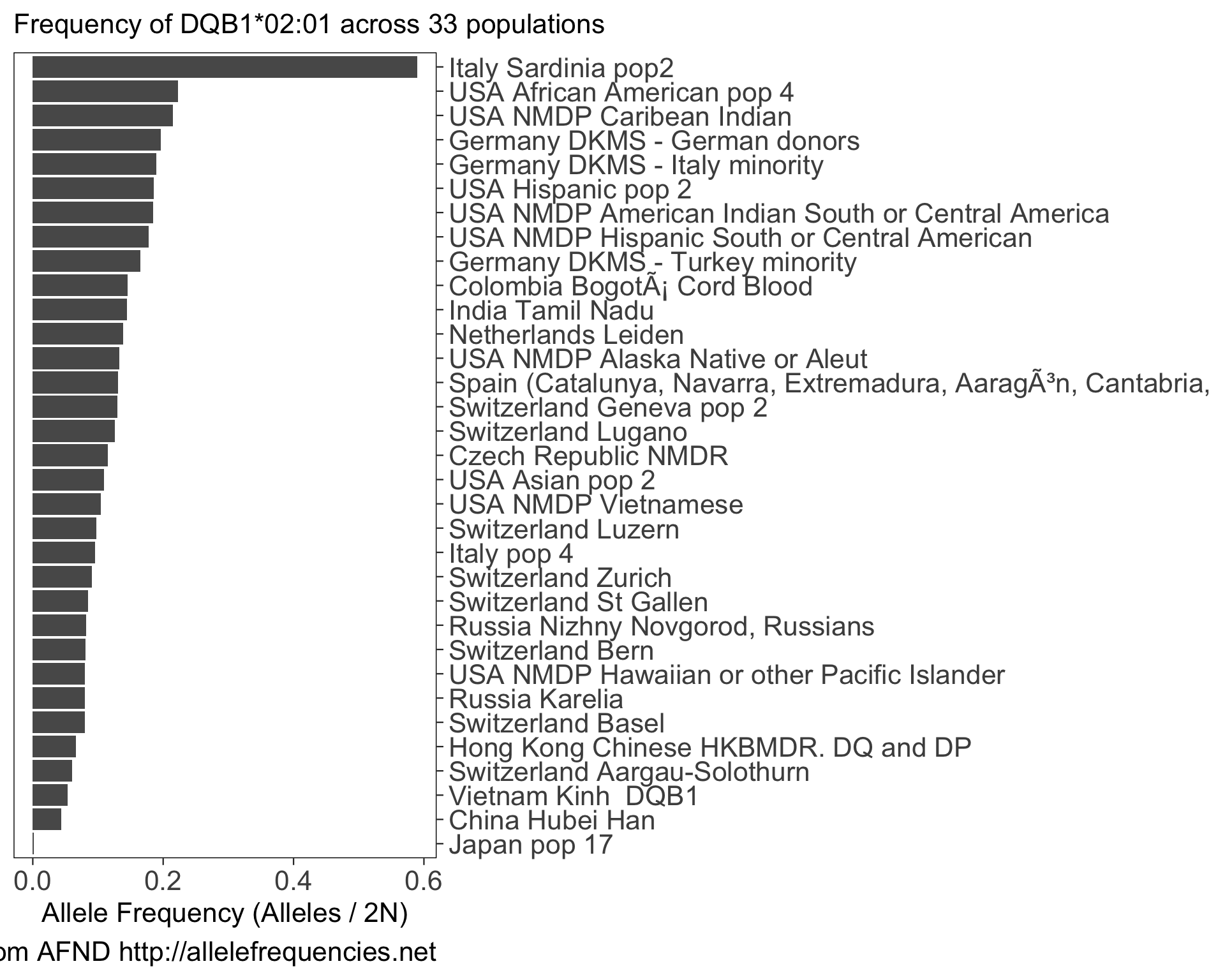
hlabud provides methods to retrieve sequence alignment data from IMGTHLA and convert the data into convenient R matrices ready for downstream analysis. See the usage examples to learn how to use the data with logistic regression and dimensionality reduction.
For example, let’s consider a simple question about two HLA genotypes.
What amino acid positions are different between these two genotypes?
library(hlabud)
a <- hla_alignments("DRB1")
dosage(a$onehot, c("DRB1*03:01:05", "DRB1*03:02:03"))
## F26 Y26 D28 E28 F47 Y47 G86 V86
## DRB1*03:01:05 0 1 1 0 1 0 0 1
## DRB1*03:02:03 1 0 0 1 0 1 1 0From this output, we can conclude that four positions (26, 28, 47, 86) distinguish these two HLA-DRB1 alleles. We see that DRB1*03:01:05 has a Y at position 26 and DRB1*03:02:03 has a F.
Examples
See the usage examples to get some ideas for how to use hlabud in your analyses.
Citation
hlabud provides access to the data in IMGT/HLA database. Therefore, if you use hlabud then please cite the IMGT/HLA paper:
- Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48: D948–D955. doi:10.1093/nar/gkz950
hlabud also provides access to the data in Allele Frequency Net Database (AFND). Therefore, if you use hlabud::hla_frequencies() then please cite the AFND paper:
- Gonzalez-Galarza FF, McCabe A, Santos EJMD, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48: D783–D788. doi:10.1093/nar/gkz1029
Additionally, you can also cite the hlabud package like this:
- Slowikowski K. hlabud: methods for access and analysis of the human leukocyte antigen (HLA) gene sequence alignments from IMGT/HLA. R package version 1.0.0.
Related work
I recommend this article for anyone new to HLA, because the beautiful figures help to build intuition:
- La Gruta NL, Gras S, Daley SR, Thomas PG, Rossjohn J. Understanding the drivers of MHC restriction of T cell receptors. Nat Rev Immunol. 2018;18: 467–478.
Learn about the conventions for HLA nomenclature:
- Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75: 291–455.
For case-control analysis of HLA genotype data, consider the BIGDAWG R package available on CRAN. Here is the related article:
- Pappas DJ, Marin W, Hollenbach JA, Mack SJ. Bridging ImmunoGenomic Data Analysis Workflow Gaps (BIGDAWG): An integrated case-control analysis pipeline. Hum Immunol. 2016;77: 283–287.